Challenge:
To identify new compounds in the treatment regimen of candidiasis by implementing a novel approach to screen the targets of FDA-approved drugs for structural homology with Candida proteome, followed by ligand fingerprint-based virtual screening for the identification of novel
inhibitors.
Solution:
BIOVIA Discovery studio and Pipeline Pilot
Benefits:
- Target and ligand fingerprint‐based drug
screening - High‐precision molecular docking
- Novel and systematic investigation
- Virtual screening and invitro testing
CHALLENGE: Untangling a novel approach to screen and identify leads candidates for candidiasis
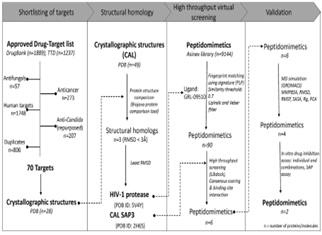
Fig 1:The workflow for identification of novel targets and drugs for Candida management
To combat this scientific issue, the customer has devised a novel strategy for funnelling inhibitors as depicted in the Fig 1. Nevertheless, their perspective on drug target was relatively interesting to investigate the Structural homology and binding site comparison of Candida albicans (CAL) secreted aspartyl proteinase (SAP3) with HIV‐1 protease using Biojava protein comparison tool drives their research more meticulously. Finally, the virtually funnelled compounds are probed for docking and molecular dynamics simulation as a cost effective technique to prioritize the molecules for in vitro testing.
RESULT: Experimental correlation with the computational study
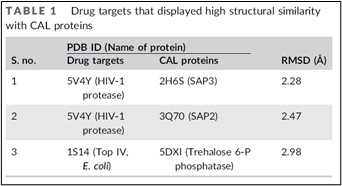
This client has tapered FDA-approved drug targets with various filter parameters them using BIOVIA Pipeline Pilot 17.2 (BIOVIA, 2020a) to obtain 28 crystallographic structures in Protein Data Bank. The structural and binding site similarities of these 28 drug targets were compared to the 49 unique structures of CAL proteins available in PDB. Among, only three drug targets displayed RMSD < 3 Å with CAL proteins(Table 1)
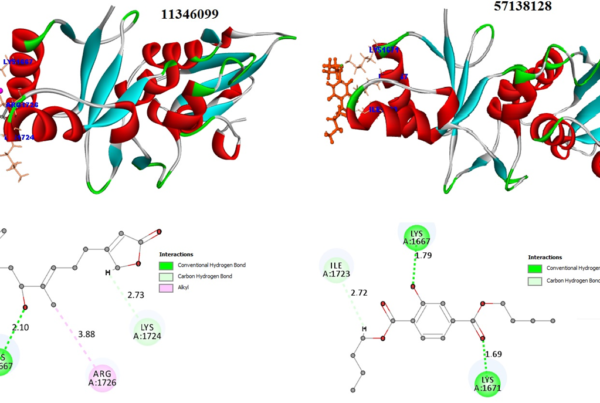
Both proteins belong to the aspartyl protease family. Structural analysis of catalytic sites revealed that Asp32 of CAL SAP3, a critical residue for enzyme activity of aspartyl proteases, is conserved and corresponded to Asp25 of HIV‐1 protease Fig 2. As inferred from the structure‐based sequence alignment.
Molecular docking of GRL‐09510 to HIV‐1 protease and CAL SAP3 CDOCKER energy scores are highly comparable at the active sites of HIV‐1 protease (−46.2 kcal/mol) and CAL SAP3 (−46.8 kcal/mol) bolster the study to use GRL‐09510 as a reference molecule to screen the database of 9144 peptidomimetic compounds using ligand fingerprints approach of Tanimoto similarity index and oral bioavailibity.
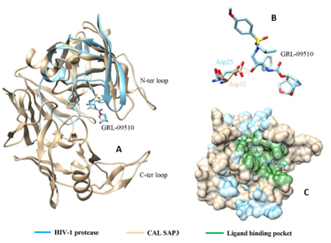
Fig 2: (A): Superimposed 3D structures of HIV‐1 protease (5V4Y, in blue) and CAL SAP3 (2H6S, in tan). The inhibitor GRL‐09510 is represented as stick model in the binding site; (B): H‐bond interaction of GRL‐09510 with residue Asp25 of HIV‐1 protease, and superimposed Asp32 of CAL SAP3; (C): The superimposed binding site surface (in green) of HIV‐1 protease and CAL SAP3 with GRL‐09510.
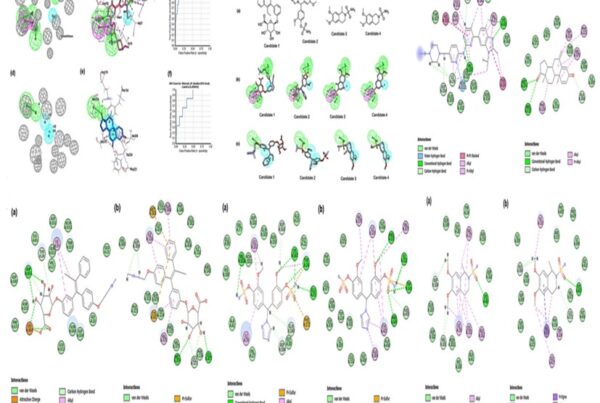
Further based on docking, interaction of six peptidomimetics was found with important catalytic residues Asp32 and/or Asp218 of SAP3, similar to the interaction of a known protease inhibitor (pepstatin A). Of six peptidomimetics, MD simulations stability was higher for P12, P21, P104, and P124 peptidomimetics and pepstatin A (control) as compared to P143 and P158 bound structures based on RMSD, Rg (radius of gyration), solvent accessible surface area (SASA), and root mean square fluctuation (RMSF) plots. Also, P21 and P124 showed significant inhibition of SAP at pH 3.5, as reflected in the decrease in absorbance and protease activity. P21 and P124 when used in combination with fluconazole have shown to reduce the MIC dose of fluconazole required to arrest the growth of CAL. Besides, the binding of P21 and P124 to the isozymes secreted aspartyl protease (SAP 1-3) is correlated with their interaction to Asp32 and Asp218. The results of their research may therefore be useful in the development of novel aspartyl protease inhibitors that can fight Candida infections efficiently.
SOLUTION: Nutshell approach of bio via solutions with other tools will mitigate the Candida infections
To address the issue of MDR issues associated with commercially accessible medications, this customer conducted creative research using BIOVIA’s Pipeline Pilot and Discovery Studio. This customer used a cutting-edge, a methodical investigative technique that used high-precision molecular docking to resolve the targeted protein and ligands fingerprint-based drug screening.
There are currently no drugs available or undergoing clinical trials for the Candida SAPs. Previously, it was reported that CAL biofilm inhibitory effect when HIV aspartyl protease inhibitors,nelfinavir, ritonavir, and saquinavir were used in combination with caspofungin or amphotericin B than when present alone; these inhibitors were not as effective in combination with fluconazole. Hence, Combinatorial drug therapy that targets diverse components of the fungal cell is an apt strategy to evade or delay drug resistance
In a nutshell, This customer identified two novel peptidomimetics (P21 and P124) that are structurally diverse and functionally equivalent to pepstatin A as a anti‐Candida compounds that can function as SAP inhibitors to control the growth of Candida and highlights the utility of target and ligand fingerprint‐based drug screening for expanding the antifungal drug space. The clinical efficacy and safety of these peptidomimetics have not been addressed in this study
Cite: Chakraborty, S., Rahate, K., Kumar, C., & Idicula‐Thomas, S. (2022). Expanding the therapeutic options for Candida infections using novel inhibitors of secreted aspartyl proteases. Drug Development Research, 1–14. https://doi.org/10.1002/ddr.22015
Acknowledgement: We, team thank Dr. Susan Idicula Thomas, Scientist-E, Biomedical Informatics Centre, National Institute for Research in Reproductive and Child Health, Mumbai, India for sharing their one of the research publication as a case study.
Publication Date : January, 2023