CHALLENGE: Dual Inhibitors against CDK4/6 and Aromatase Enzyme
This client utilized the BIOVIA Discovery studio suite, Discovery Studio 2019 (DS.v.2019) to design the dual inhibitors for cancer.
They have used “prepare protein” tool under macromolecule module for the preparation of selected proteins PDB’s (5L2S and 3S7S) to construct structure-based pharmacophore (SBP) model. Further, The CDOCKER protocol based on simulation annealing and CHARMm in DS.v.2019 was used to perform molecular docking and validated in terms of RMSD to ensure the accuracy of the docking protocol.
Further, Discovery Studio Visualizer 2019 was used to investigate the protein–ligand interaction complexes. Also, insilico pharmacokinetics to predict adsorption, distribution, metabolism, excretion, and toxicity (ADMET) parameters for the identified dual inhibitor candidates using ADMET descriptors and toxicity prediction TOPKAT tools.
The defined ADMET descriptors, such as HIA(Human intestinal absorption), calculated at aqueous solubility at 25 °C, BBB(Blood brain barrier) penetration after oral administration, PPB(Plasma protein binding), CYP2D6 inhibition, and hepatotoxicity, were evaluated for the selected candidates will broaden the likelihood for future drug-molecule development.
Nevertheless, calculations based on DFT were utilized to determine a compound’s primary molecular properties. In this research, DFT calculations were used to optimize the 3D structures of the four identified candidates. These structures were subsequently used in the computations of the molecular orbitals, that is, the highest occupied molecular orbital (HOMO), lowest unoccupied molecular orbital (LUMO), and energy gap (ΔE).
RESULT: Structure-based pharmacophore model for screening and identification of dual inhibitors with lead optimization
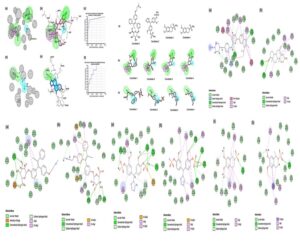
The Structure-based pharamcophore model was constructed for co-crystal ligands abemaciclib (PDB ID: 5L2S) for CDK6 and exemestane (PDB ID: 3S7S) for aromatase with ROC validation. The model only with good ROC (receiver operating characteristic curve) was selected with a good selectivity score for screening. The model with pharmacophore feature (AAAADH) of CDK6 protein and aromatase (AHHH) selected based on validation was used for screening of compounds. In total of 755 compounds (37 and 718 from CDK6 and aromatase) were obtained based on fit value with two Structure-based pharmacophore model.
The final compounds were selected in common fit with both SBP and its CDOCKER negative energy values are favourable, shows potential binding and its interaction was depicted in the Fig 1.
Fig 1: Representation of pharmacophore models and protein-ligand interactions
These screened candidates are further funnelled for pharmacokinetics properties and the results are interpreted based on the graphical representation plotted against ADMET_AlogP98 vs. ADMET_PSA 2D (Fig 2) in the optimum prediction space (OPS), it shows a confidence, level of 95% and 99% for BBB and human intestinal absorption, respectively. The compounds within OPS shows compounds confidence with ADMET properties
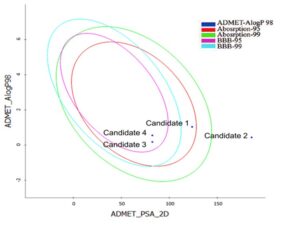
Fig 2: ADMET profiling of ligand moleculesSOLUTION: Discovery Studio-suite is a complete solution for drug discovery and development
CDK4/6 and aromatase biomarkers are frequently diagnosed in breast cancer, the concurrent inhibition of CDK4/6 and aromatase benefits the therapeutic potential and treatment. The proposed strategy will help to develop a dual-target inhibitor.
Currently, there is no evidence using computational method reported in the identification of dual inhibitors of CDK4/6 and aromatase. To achieve the design of dual inhibitors, a virtual screening workflow was applied, including the generation of pharmacophore models, followed by pharmacophore-based virtual screening of seven thousand compounds from the sc-PDB database and molecular docking studies of identified candidates, MD (Molecular dynamics) simulation, and ADMET analysis bolster the compound properties.
Based on these observations, four potential candidates could be possible dual inhibitors of CDK4/6 and aromatase. Hence, further validation using invitro, invivo cell-based activity and synthesis of these candidate for CDK4/6-aromatase dual inhibitors is under progress to prove the potential of the compounds.
Acknowledgment
We, thank H. Yogish Kumar and his team, Department of Pharmaceutical Chemistry at JSS College of Pharmacy, Mysuru for providing their research case study.
To cite: Adon, T.; Shanmugarajan, D.; Ather, H.; Ansari, S.M.A.; Hani, U.; Madhunapantula, S.V.; Honnavalli, Y.K. Virtual Screening for Identification of Dual Inhibitors against CDK4/6 and Aromatase Enzyme. Molecules 2023, 28, 2490.
Publication Date : March, 2023